SAS Thread - esasmosaic - XMM-Newton
Creation of EPIC merged background subtracted and exposure corrected images
|
Introduction This thread describes how to create EPIC merged background subtracted and exposure corrected images combining data from three ObsIDs and the three instruments. (This thread used to describe the creation of a merged source list. That capability has issues in the current SAS, but will return with the next version.) This thread uses data from three observations of M101: ObsIDs 0104260101, 0164560701, and 0212480201. These data are also used as the Mosaicking Analysis example where a script and output files are provided. Expected Outcome The final outcome of this thread are images of the merged data of M101. SAS Tasks to be Used Prerequisites This thread starts with the assumption that data from the three EPIC instruments from the three individual observations have been processed following the Image Analysis thread, with the slight complication of processing three energy bands: 0.4-0.75 keV, 0.75-1.3 keV, and 2.0-7.2 keV. SAS must be active and the $SAS_CCF environment variable should point to the ccf.cif file from one of the obsevations. Useful Links
Last Reviewed: 31 January 2025, for SAS v22.0Last Updated: 25 May 2023 |
Procedure
This thread contains a step-by-step recipe to create EPIC background subtracted and exposure corrected images from merged observations of the nearby galaxy M101. Note that this thread is meant for observations taken of similar parts of the sky, and not necessarily mosaic mode observations.
- Process data from the three observations and three detectors following the Image Analysis thread in the three energy bands: 0.4-0.75 keV, 0.75-1.3 keV, and 2.0-7.2 keV. For convenience, use the directory structure (for example),
/M101/merge
/M101/0104260101/proc
/M101/0104260101/odf
/M101/0164560701/proc
/M101/0164560701/odf
/M101/0212480201/proc
/M101/0212480201/odfwhere /M101/merge is the directory for the merged image processing, the proc directories contain the individual ObsID image processed data, and the odf directories contain the individual ObsID ODF files. SAS must be active and the $SAS_CCF environment variable should point to the ccf.cif file from one of the observations, for example,
/M101/0104260101/proc/ccf.cif - Create an ASCII file which contains paths to the individual observation exposures (mosaic.dat). They can be the relative paths.
mosaic.dat
../0104260101/proc/mos1S001
../0104260101/proc/mos2S006
../0104260101/proc/pnS005
../0164560701/proc/mos1S001
../0164560701/proc/mos2S002
../0164560701/proc/pnS003
../0212480201/proc/mos1S001
../0212480201/proc/mos2S002
../0212480201/proc/pnS003 - Merge the individual components (count, exposure, particle background, and soft proton images) from the three observations, three instruments, and three bands. The first time will include point sources. Note that type parameter is 1 for the count image, 2 for the exposure map, 3 for the quiescent particle background, 4 for the soft proton flare image, and 5 for the SWCX image. type=7 is for merging the cheese maps.
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=400 \
ehigh=750 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=400 \
ehigh=750 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=400 \
ehigh=750 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=400 \
ehigh=750 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=400 \
ehigh=750 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=750 \
ehigh=1300 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=750 \
ehigh=1300 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=750 \
ehigh=1300 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=750 \
ehigh=1300 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=750 \
ehigh=1300 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=2000 \
ehigh=7200 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=2000 \
ehigh=7200 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=2000 \
ehigh=7200 withcheese=no xdim=1100 ydim=1100 alpha=1.7
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=2000 \
ehigh=7200 withcheese=no xdim=1100 ydim=1100 alpha=1.7<
mosaicmerge dirfile=mosaic.dat coordsys=2 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=2000 \
ehigh=7200 withcheese=no xdim=1100 ydim=1100 alpha=1.7 - Create background subtracted, exposure corrected, and adaptively smoothed images in the three individual bands and a combined soft band.
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=400 \
ehighlist=750 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes \
withmaskcontrol=no
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=750 \
ehighlist=1300 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes \
withmaskcontrol=no
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist='400 750' \
ehighlist='750 1300' withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes \
withmaskcontrol=no
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=2000 \
ehighlist=7200 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes \
withmaskcontrol=noRename the files.
mv merged-rate-400-750.fits merged-rate-400-750-ps.fits
mv merged-rate-750-1300.fits merged-rate-750-1300-ps.fits
mv merged-rate-400-1300.fits merged-rate-400-1300-ps.fits
mv merged-rate-2000-7200.fits merged-rate-2000-7200-ps.fitsThen display the results.
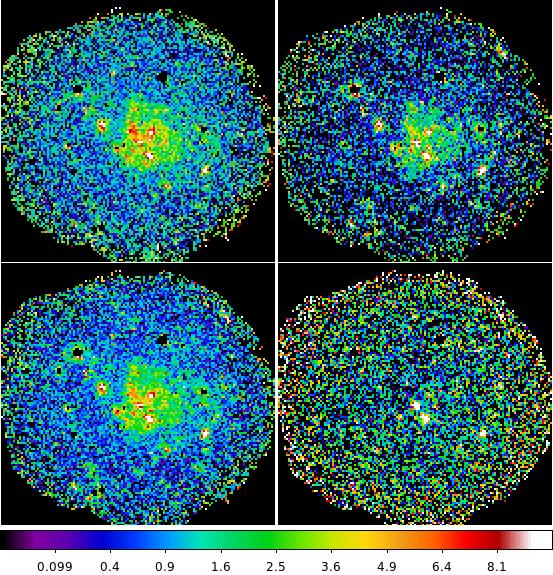
ds9 merged-rate-400-750-ps.fits merged-rate-750-1300-ps.fits merged-rate-400-1300-ps.fits \
merged-rate-2000-7200-ps.fits & - Next create images which exclude point sources using the cheese masks for the individual observations.
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=6 elow=400 \
ehigh=750 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=6 elow=750 \
ehigh=1300 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=1 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=2 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=3 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=4 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=5 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100
mosaicmerge dirfile=mosaic.dat coordsys=2 alpha=1.7 \
crval1=210.85 crval2=54.35 pixelsize=0.03 type=6 elow=2000 \
ehigh=7200 withcheese=true cheesemasktype=t xdim=1100 ydim=1100Create background subtracted, exposure corrected, and adaptively smoothed (and binned) images in the three individual bands and a combined soft band.
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=400 \
ehighlist=750 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes \
withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=750 \
ehighlist=1300 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes\
withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist='400 750' \
ehighlist='750 1300' withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes\
withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=true smoothingcounts=50 maskthresh=0.02 elowlist=2000 \
ehighlist=7200 withbinning=true binning=2 withpartbkg=yes withspbkg=yes withswcxbkg=yes\
withmask=true mask=mosaic-cheeset.fitsRename the files.
mv merge-rate-400-750.fits merge-rate-400-750-nps.fits
mv merge-rate-750-1300.fits merge-rate-750-1300-nps.fits
mv merge-rate-400-1300.fits merge-rate-400-1300-nps.fits
mv merge-rate-2000-7200.fits merge-rate-2000-7200-nps.fitsThen display the results.
ds9 merge-rate-400-750-nps.fits merge-rate-750-1300-nps.fits merge-rate-400-1300-nps.fits \
merge-rate-2000-7200-nps.fits & - Make simple binned images.
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist=400 \
ehighlist=750 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmaskcontrol=no
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist=750 \
ehighlist=1300 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmaskcontrol=no
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist='400 750' \
ehighlist='750 1300' withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmaskcontrol=no
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist=2000 \
ehighlist=7200 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmaskcontrol=nobinadaptmerge withsmoothing=false maskthresh=0.02 elowlist=400 \
ehighlist=750 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist=750 \
ehighlist=1300 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist='400 750' \
ehighlist='750 1300' withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmask=true mask=mosaic-cheeset.fits
binadaptmerge withsmoothing=false maskthresh=0.02 elowlist=2000 \
ehighlist=7200 withbinning=true binning=2 withpartbkg=yes withspbkg=yes \
withswcx=yes withmask=true mask=mosaic-cheeset.fitsThen display the results.
ds9 bin-400-750.fits bin-750-1300.fits bin-400-1300.fits bin-2000-7200.fits &
|
Caveats |
- Removed a total of (1) style text-align:center;
- Removed a total of (17) style text-align:justify;
- Removed a total of (2) border attribute.
- Converted a total of (5) center to div.








































 Sign in
Sign in
 Science & Technology
Science & Technology